Rapid Processing of Casework and Reference DNA Samples Using Standard Protocols
1Promega UK, 2Forensic Science Ireland, Garda Headquarters, Eire
Publication Date: 2015
Introduction
The need to reduce processing time for forensic DNA samples has grown in the last few years. Laboratories must increase efficiencies to minimise backlogs and reduce costs. A number of rapid DNA solutions were recently brought to market, mostly for point-of-sampling DNA analysis, and are designed primarily for processing reference samples (1) (2) . While these solutions may yet be proven as viable and valuable options for point-of-sampling applications, the relatively high instrumentation and per-sample costs are prohibitive for established forensic DNA laboratories, which have invested heavily in other laboratory technologies and staff training. A more feasible approach to increase both laboratory efficiency and the work capacity of existing equipment in such laboratories might be to adopt rapid-cycling protocols and, for database samples, direct amplification. Direct-amplification technologies from Promega Corporation, Life Technologies and most recently Qiagen eliminate the need for expensive and time-consuming DNA extraction procedures (3) . These technologies have reduced amplification time from approximately 3 hours to around 45 minutes, depending on kit and protocol optimisations.
A rapid solution for casework was not available until the release of the PowerPlex® ESX Fast and ESI Fast Systems. These kits amplify casework samples in approximately 50 minutes (4) . The time savings from the amplification alone can significantly increase both laboratory efficiency and work capacity of existing equipment, reducing the requirement for capital expenditure should sample numbers increase.
In this article, we demonstrate the use of the PowerPlex® ESX 17 Fast System to rapidly process casework and reference DNA samples in an operational forensic DNA laboratory using the manufacturer’s recommended protocols (5) . Reference samples included buccal cells on FTA® cards, OmniSwab™ buccal scrapes and cotton swab buccal scrapes. FTA® samples were amplified directly in half-volume (12.5µl) and full-volume (25µl) reactions. Swabs were pretreated with SwabSolution™ Reagent prior to amplification. The total time from sample to analysis for 21 FTA® card punches, in duplicate, or 21 buccal swabs (including time to process swabs with SwabSolution™ Reagent) and PCR controls was 2 hours and 30 minutes. Casework samples were chosen from a batch processed the previous day using the laboratory’s standard procedures. Amplification required approximately 55 minutes.
Processes were not optimised prior to these experiments, and the PowerPlex® ESX 17 Fast System had not been previously used in this laboratory. Despite this, high-quality results were obtained from all reference and casework samples in both full-volume (25µl) and half-volume (12.5µl) reactions. This work illustrates the possibility for rapid, high-quality processing of all types of forensic DNA samples in established laboratories using off-the-shelf technologies and protocols.
Materials and Methods
Buccal cells on FTA® cards: We chose 21 buccal samples on FTA® cards that were previously created by Forensic Science Ireland (FSI) staff and processed numerous times. These cards provided 14 full profiles and 7 partial profiles with the existing validated STR system (AmpFlSTR® NGM SElect™ Kit). FTA® cards were punched using a BSD Duet automated puncher using a 1.2mm punching head. PowerPlex® ESX 17 Fast PCR amplification mix was prepared according to the PowerPlex® ESX 17 Fast System Technical Manual #TMD040 (5) . Direct-amplification reactions were performed in full-volume (25µl) and half-volume (12.5µl) reactions on an Arktik™ thermal cycler (Thermo Scientific). As no specific information about the composition of the block could be found, we took a “worst case” approach and modified the thermal cycling protocol slightly (Table 1) to try to account for the reduced efficiencies of aluminium blocks. Sample mode was selected, and cycling was carried out for 26 cycles according to the kit manufacturer’s recommendation for initial optimisation experiments.
Buccal swabs: We collected 21 freshly prepared buccal swabs (11 Whatman OmniSwab™ and 10 cotton swabs) from FSI staff and incubated each swab in a 1.5ml microcentrifuge tube with 1ml of SwabSolution™ Reagent for 30 minutes at 70°C on a Thermo-Block TBD120 static heat block.
We then transferred 2µl of SwabSolution™ extract from each sample directly to 23µl of PowerPlex® ESX 17 Fast PCR amplification mix as directed in the PowerPlex® ESX 17 Fast System Technical Manual. All swab amplifications were carried out at full-reaction volume (25µl). We did not evaluate half-volume reactions because the ability of the PowerPlex® ESX 17 Fast System to amplify SwabSolution™ extracts in half-volume reactions was previously demonstrated (3) . Samples were amplified using an Arktik™ thermal cycler for 26 cycles using the modified protocol described in Table 1. To compare the performance of the two cycling modes of the Arktik™ thermal cycler, we amplified one set of samples using Sample mode and the other using Block mode.
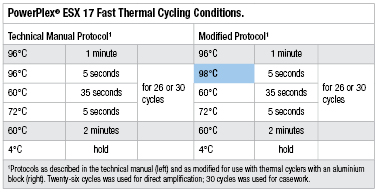 Table 1. PowerPlex® ESX 17 Fast Thermal Cycling.
Table 1. PowerPlex® ESX 17 Fast Thermal Cycling. Casework samples: As it was not possible to perform a direct back-to-back comparison of the PowerPlex® ESX 17 Fast System and NGM SElect™ Kit using live casework samples, we chose 19 previously analysed casework samples that had been extracted using the QIAsymphony® DNA Investigator kit (Qiagen) on a Hamilton STAR robotic platform. We included some samples that did not reach the recommended 1ng of template DNA in the AmpFlSTR® NGM SElect™ reactions as well as some samples that did. These samples were quantified previously using Quantifiler® Duo on an Applied Biosystems 7500 Real-Time PCR System. Samples were amplified using the AmpFlSTR® NGM SElect™ Kit at full-reaction volume (25µl) with up to 1ng of template DNA (maximum of 10µl sample volume) for 29 cycles, according to the kit manufacturer’s recommended protocols (6) , on the Arktik™ thermal cyclers using Sample mode.
Samples were re-amplified using the PowerPlex® ESX 17 Fast System, a 25µl reaction volume and the modified thermal cycling protocol described in Table 1 for 30 cycles. Sample input volumes were calculated based on DNA concentrations determined using the Quantifiler® Duo Kit, with a target of 0.5ng of template DNA. For samples where the DNA concentration was too low to achieve the desired mass of target DNA, we added the maximum sample volume allowed (17.5μl) to the amplification reaction. The sample descriptions, DNA concentrations, PCR input volumes and DNA amounts are shown in Table 2.
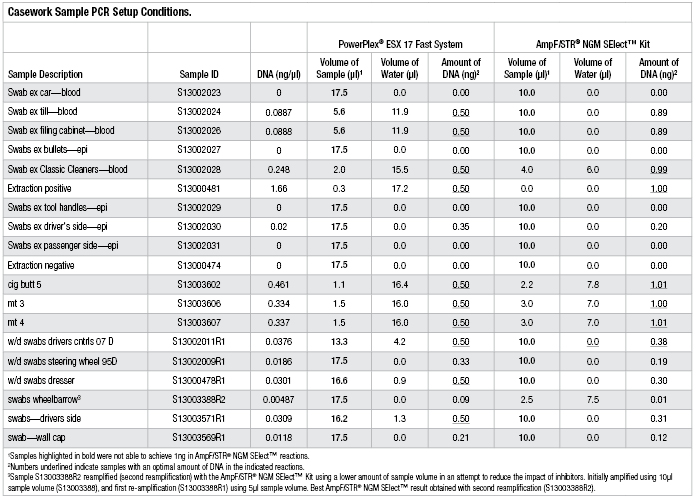 Table 2. Casework Sample PCR Setup Conditions.
Table 2. Casework Sample PCR Setup Conditions. All PCR products were prepared for electrokinetic injection according to manufacturer's instructions (5) (6) and then separated on an Applied Biosystems® 3500xL Genetic Analyzer using default run conditions for POP-4® polymer with a 36cm array. Injections were carried out for 24 seconds at 1.2kV. Data were analysed using GeneMapper® ID-X software, version 1.2.
As the PowerPlex® ESX 17 Fast System has not been validated in this laboratory, analytical and stochastic thresholds were not available. The thresholds currently in use in the laboratory (optimised with the AmpFlSTR® NGM SElect™ Kit) were used for all analyses unless otherwise stated. The AmpFlSTR® NGM SElect™ thresholds currently used by FSI are
Reference samples (3500xL):
Analytical threshold 400RFU
Stochastic threshold 560RFU
Heterozygote balance threshold 50%
Casework samples (3500xL):
Analytical threshold 175RFU
Stochastic threshold 560RFU
Heterozygote balance threshold 70%
Results
Buccal Cells on FTA® Cards
The total time taken to process 21 FTA® cards, including assembling the PCR and punching the cards in duplicate was 2 hours and 30 minutes. Of this, the thermal cycling took 55 minutes, which was 10 minutes longer than the GeneAmp® 9700 or Veriti® thermal cycler, suggesting that either the Arktik™ thermal cyclers were not ramping at 4°C/s as indicated or the Sample mode was slowing the program down.
Of the 21 FTA® cards chosen for this study, three samples were removed from the data set due to poor CE injection (one sample) and off-scale peaks (two samples). All three occurred with the 12.5µl reaction volume and PowerPlex® ESX 17 Fast System. These samples were excluded from the data set for comparison of 12.5µl and 25µl PowerPlex® ESX 17 Fast reactions and the AmpFlSTR® NGM SElect™ data set. These data were included in the peak count and peak height data.
Profile Success
Profile success was assessed using the analytical, stochastic and heterozygote balance thresholds defined above.
- The full-volume (25µl) PowerPlex® ESX 17 Fast reactions provided 12 full profiles and 6 partial profiles (15 full and 6 partial profiles if the removed samples are included).
- The half-volume (12.5µl) PowerPlex® ESX 17 Fast reactions provided 17 full profiles and 1 partial profile.
- The AmpFlSTR® NGM SElect™ samples provided 12 full profiles and 6 partial profiles (14 full and 7 partial profiles if the removed samples are included).
Peak Counts and Peak Height Comparison
The PowerPlex® ESX 17 Fast half-volume reactions tended to produce higher peak heights than the PowerPlex® ESX 17 Fast full-volume reactions and NGM SElect™ full-volume reactions (Figure 1). Example electropherograms of the same sample processed with the three methods are shown in Figure 2.
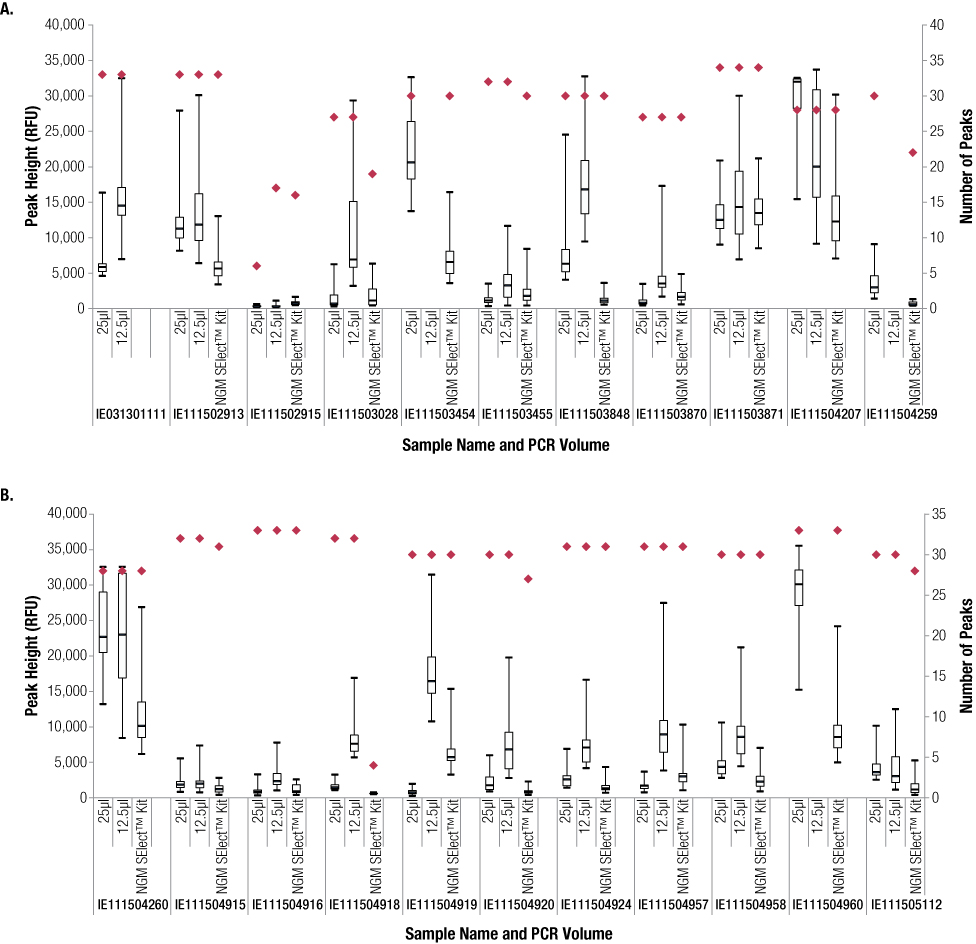 Figure 1. Comparison of the number of peaks and peak height for full- and half-volume PowerPlex® ESX 17 Fast and NGM SElect™ FTA® samples.
Figure 1. Comparison of the number of peaks and peak height for full- and half-volume PowerPlex® ESX 17 Fast and NGM SElect™ FTA® samples. Red diamonds indicate number of peaks. Boxes show peak height inter-quartile range, with the median indicated by a dash. Whiskers show minimum and maximum peak heights observed. Sample IE031301111 is a positive control, not shown for the NGM SElect™ data.
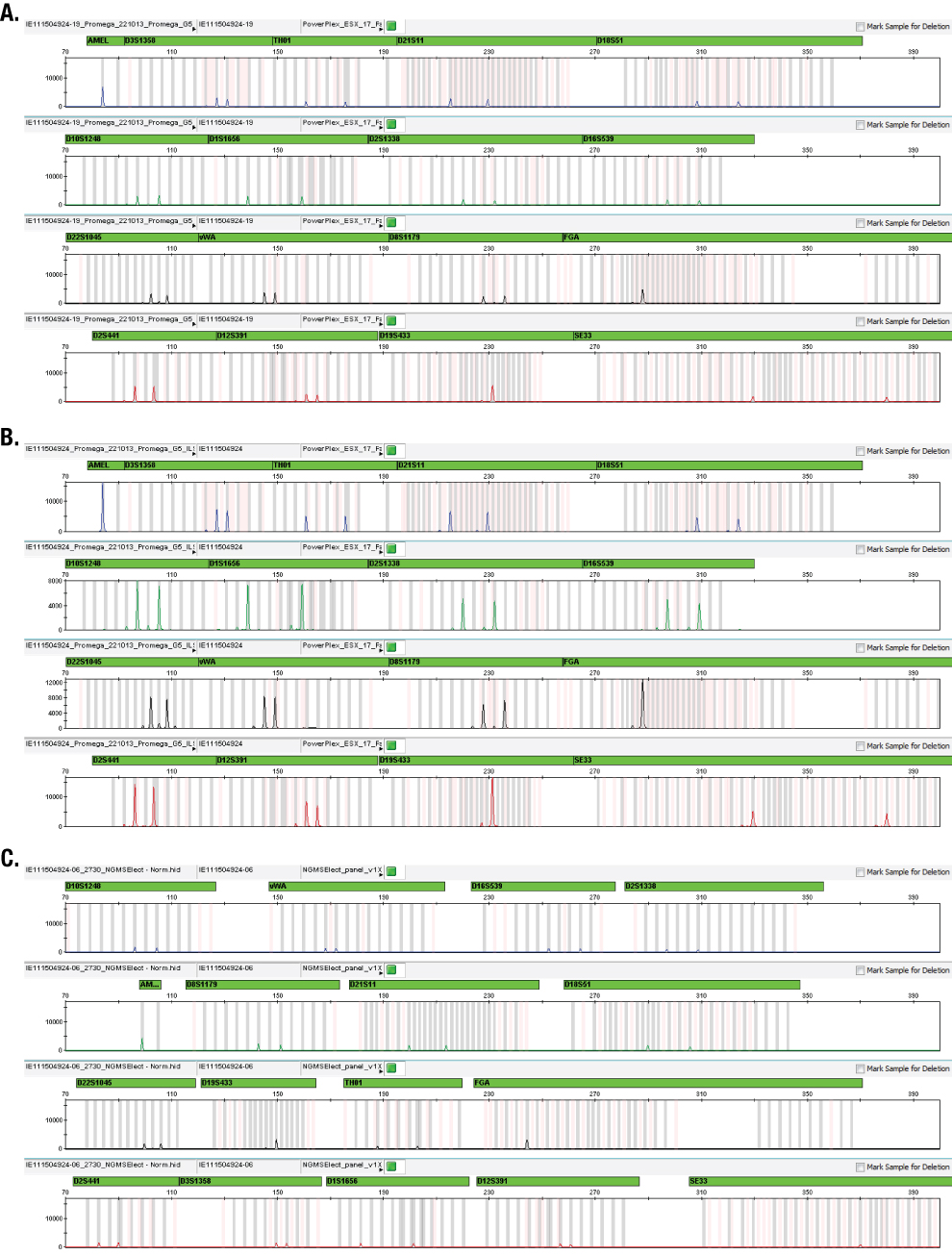 Figure 2. Example electropherograms for an FTA® sample amplified directly with the PowerPlex® ESX 17 Fast System at 25μl (Panel A), 12.5μl (Panel B) and NGM SElect™ Kit (Panel C).
Figure 2. Example electropherograms for an FTA® sample amplified directly with the PowerPlex® ESX 17 Fast System at 25μl (Panel A), 12.5μl (Panel B) and NGM SElect™ Kit (Panel C). The PowerPlex® ESX 17 Fast half-volume reactions contained eight off-scale samples (samples where one or more pink, off-scale markers were seen in GeneMapper® ID-X software) compared to four in the PowerPlex® ESX 17 Fast full-volume reactions and one in the NGM SElect™ data set. This difference between the full- and half-volume PowerPlex® ESX 17 Fast reactions was expected due to the higher DNA concentration in the half-volume reactions. These data suggest that one PCR cycle less may be more appropriate for half-volume PowerPlex® ESX 17 Fast reactions with these samples.
Buccal Swabs
All samples provided well balanced, full profiles. When sample data were compared, Sample mode appeared to provide slightly higher peaks than Block mode, and the OmniSwabs™ swabs appeared to provide slightly higher peaks than cotton swabs (Figure 3). In neither case were the differences significant.
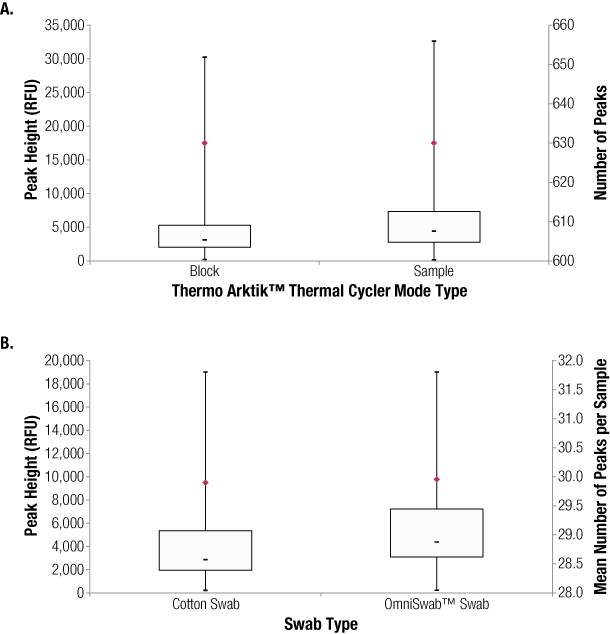 Figure 3. Results of swab extracts amplified using Block mode versus Sample mode (Panel A) and cotton swabs versus OmniSwabs™ (Panel B).
Figure 3. Results of swab extracts amplified using Block mode versus Sample mode (Panel A) and cotton swabs versus OmniSwabs™ (Panel B). Figure 4 shows example electropherograms of the same sample amplified using Block and Sample modes.
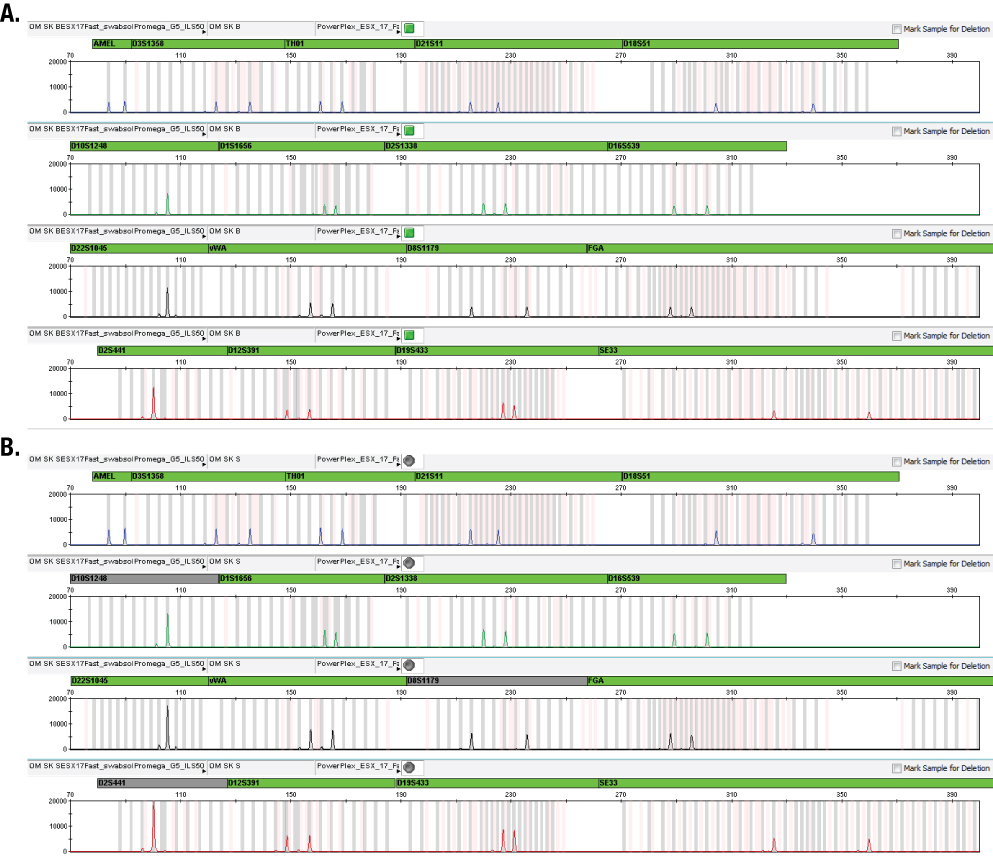 Figure 4. Example electropherograms of a swab sample processed with SwabSolution™ Reagent and amplified using the PowerPlex® ESX 17 Fast System and the Arktik™ thermal cycler using Block mode (Panel A) and Sample mode (Panel B).
Figure 4. Example electropherograms of a swab sample processed with SwabSolution™ Reagent and amplified using the PowerPlex® ESX 17 Fast System and the Arktik™ thermal cycler using Block mode (Panel A) and Sample mode (Panel B). Compared to the Block mode, the Sample mode introduced an extra ~8-second hold once the block reached the next programmed temperature and before it started the hold countdown (heating and cooling). This accounted for an extra 5 minutes in the total cycling time over the 26 cycles compared to the Block mode. The 49-minute run time with Block mode suggested that the instruments did not ramp quite as fast as the GeneAmp® 9700 PCR Systems in Max mode (~45-minute total cycling time expected on the GeneAmp® 9700 with Max ramp mode).
Casework samples
We compared the number of peaks above the analytical threshold and their respective peak heights for casework samples amplified using the AmpFlSTR® NGM SElect™ and PowerPlex® ESX 17 Fast Systems. The two kits provided roughly equivalent results for both parameters (Figure 5). However, the PowerPlex® ESX 17 Fast System tended to provide more peaks than the AmpFlSTR® NGM SElect™ Kit for samples that did not receive the optimal DNA input amount (1ng) in the NGM SElect™ Kit. The larger sample input volume possible with PowerPlex® ESX 17 Fast (17.5µl compared to 10µl in NGM SElect™ reactions), along with the fact that the kit is designed to provide balanced profiles from 0.5ng of sample DNA with 30 cycles, provided a higher chance of success with low-template samples.
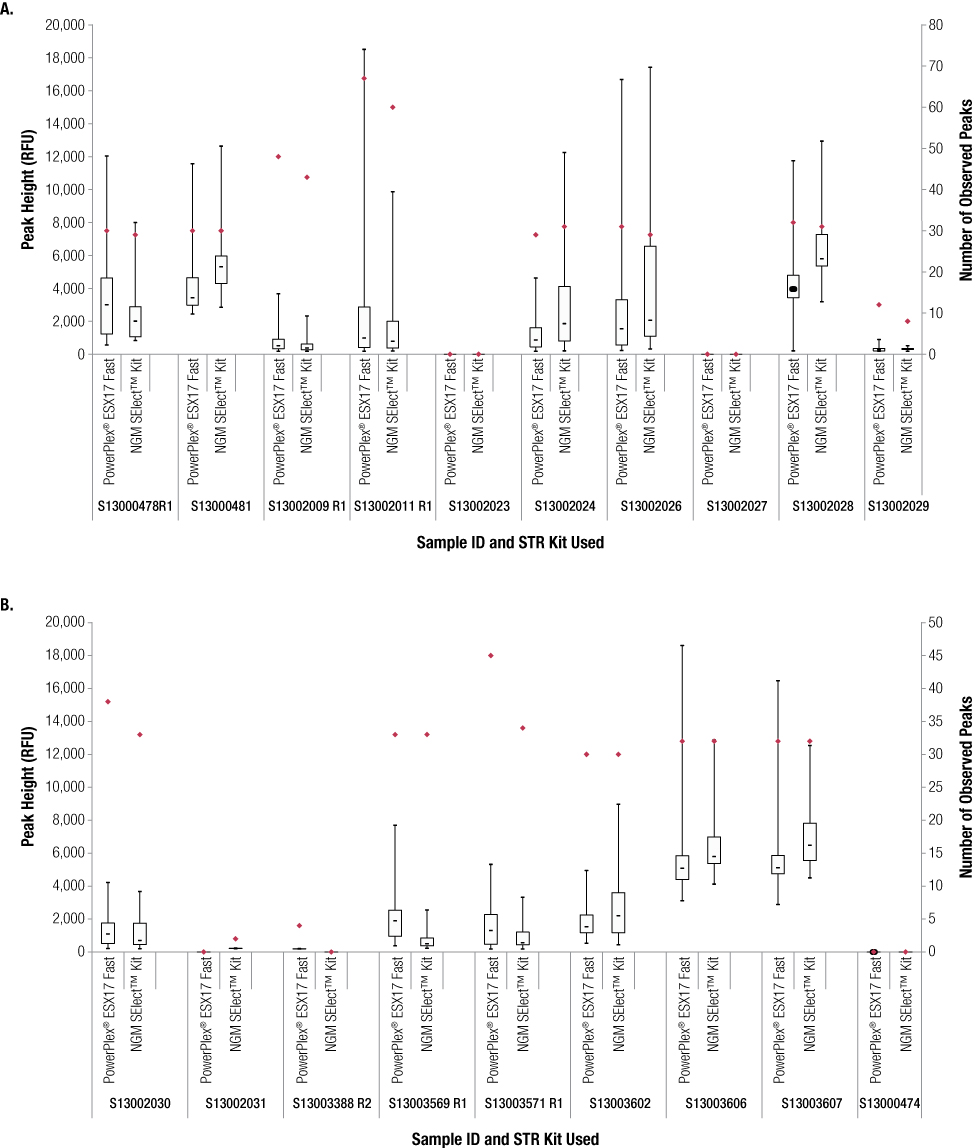 Figure 5. Comparison of the number of peaks and peak heights for the PowerPlex® ESX 17 Fast and AmpFlSTR® NGM SElect™ casework sample results.
Figure 5. Comparison of the number of peaks and peak heights for the PowerPlex® ESX 17 Fast and AmpFlSTR® NGM SElect™ casework sample results. Red diamonds indicate number of peaks. Boxes show peak height inter-quartile range, with the median indicated by a dash. Whiskers show minimum and maximum peak heights observed.
Figures 6 and 7 show example electropherograms of the same samples amplified with the PowerPlex® ESX 17 Fast and NGM SElect™ Kits and analysed using the same analysis thresholds; the differences in detected peaks are highlighted.
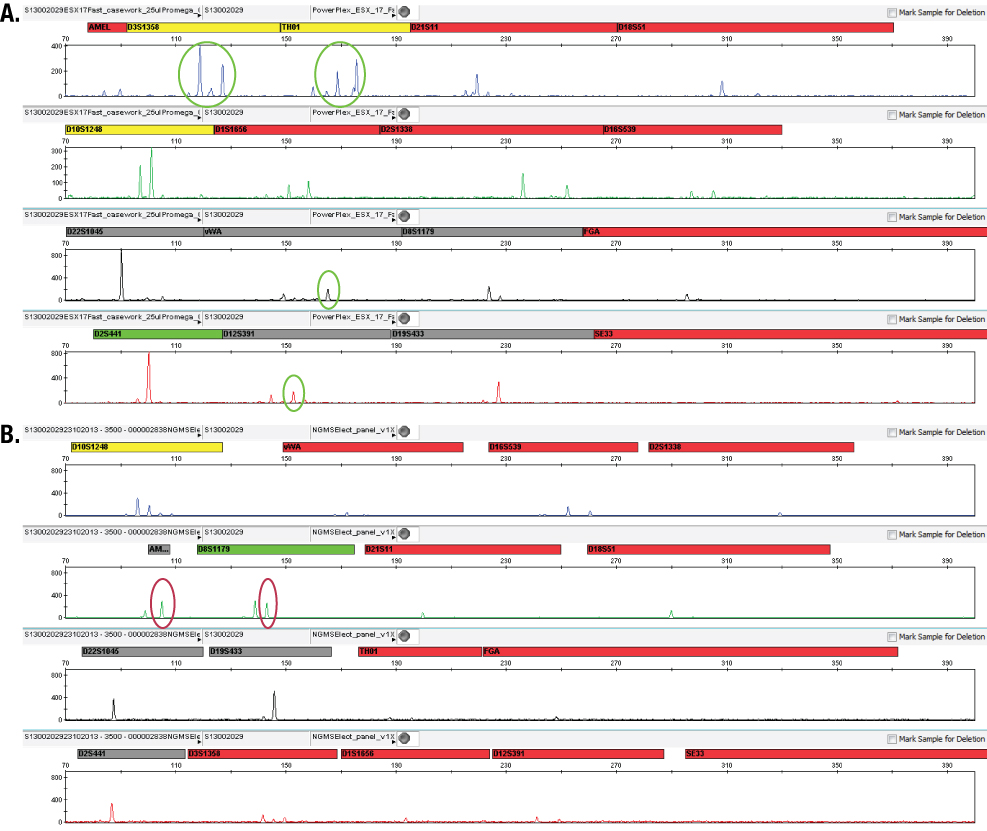 Figure 6. Electropherograms of sample S1300302029 showing different observed peaks above the analytical threshold in PowerPlex® ESX 17 Fast (Panel A) and AmpFlSTR® NGM SElect™ (Panel B) data.
Figure 6. Electropherograms of sample S1300302029 showing different observed peaks above the analytical threshold in PowerPlex® ESX 17 Fast (Panel A) and AmpFlSTR® NGM SElect™ (Panel B) data. 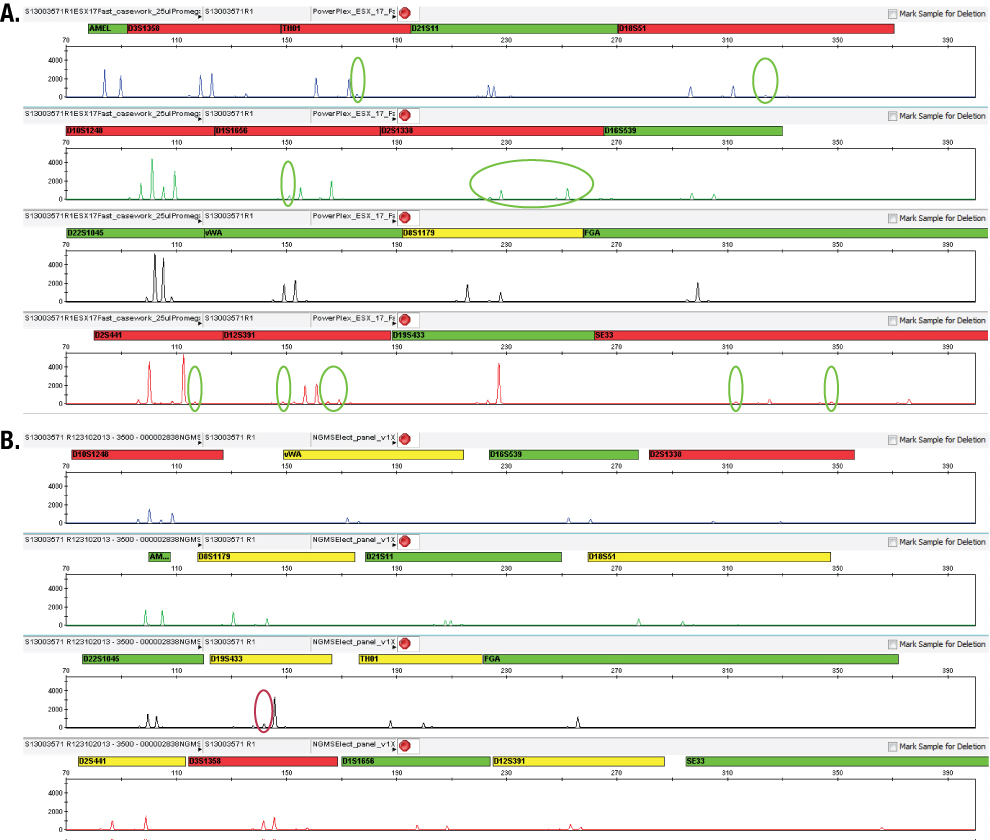 Figure 7. Electropherograms of sample S13003571 R1 showing different observed peaks above the analytical threshold in PowerPlex® ESX 17 (Panel A) Fast and AmpFlSTR® NGM SElect™ (Panel B) data.
Figure 7. Electropherograms of sample S13003571 R1 showing different observed peaks above the analytical threshold in PowerPlex® ESX 17 (Panel A) Fast and AmpFlSTR® NGM SElect™ (Panel B) data. Discussion and Conclusions
This work illustrates the possibilities for rapid workflows in established forensic DNA laboratories with no need for further capital investment. The robustness of the PowerPlex® ESX 17 Fast System and reduced processing times increased the capacity of the laboratory, while reduced-volume amplifications, along with reduced rework rates, provided significant time and cost savings.
Here we show that it is possible, in a fully equipped laboratory with trained operators using only off-the-shelf products and with no optimisation, to process up to 21 reference samples from sample to results in 2½ hours. We obtained good-quality results in all cases with both FTA® samples and swab samples.
The PowerPlex® ESX 17 Fast System compared favourably to the AmpFlSTR® NGM SElect™ Kit by providing the same, or more, full profiles from suboptimal FTA® cards when processed in full- or half-volume reactions. The increased success and robustness of reduced-volume reactions can provide significant time and cost savings for the laboratory.
The PowerPlex® ESX 17 Fast System and SwabSolution™ Reagent provided fast and robust, good-quality results from two different, common swab types.
The results obtained by amplifying extracted DNA using the PowerPlex® ESX 17 Fast System compared favourably to those of the AmpFlSTR® NGM SElect™ Kit. The PowerPlex® ESX 17 Fast System provided improved results from weak casework samples with significantly reduced PCR cycling time.
Five of the 19 casework samples achieved the optimal input DNA amount in the PowerPlex® ESX 17 Fast amplifications but not in the AmpFlSTR® NGM SElect™ reactions (Table 2). This is due to the lower optimal DNA input amount required and the larger sample volume that can be added to the PowerPlex® ESX 17 Fast reactions.
It should be noted that the AmpFlSTR® NGM SElect™ Kit also can be used with 0.5ng of input DNA and 30 cycles (6) (this protocol has not been validated at FSI and so was not assessed here). However, of the five casework samples mentioned above, three still would not have provided the optimal input DNA amount in the AmpFlSTR® NGM SElect™ reactions (S13002011R1, S13000478R1, S13003571R1). In samples with sufficient template DNA to provide full profiles, the differences in optimal DNA input amounts and maximum possible sample volumes would have limited impact. However, this does illustrate the benefits that may be achieved with low-template samples with DNA input amounts below the relative sensitivity thresholds for each kit [i.e., ≤0.0125ng/µl for AmpFlSTR® NGM SElect™ (6) and ≤0.0071ng/µl for PowerPlex® ESX 17 Fast (3) ]. The full impact of this was not assessed here as it was outside the scope of this investigation.
Acknowledgements
Thank you to Forensic Science Ireland, Dublin, for allowing us to perform these experiments as part of the demonstration of our technologies and to all their staff for their assistance and hospitality.
Thank you also to Cintia Alves at IPATIMUP for the gift of Whatman OmniSwabs™, which allowed us to include this swab type in the experiments.
Related Products
Article References
- Holland, M. and Wendt, F. (2014) Evaluation of the RapidHIT™ 200, an automated human identification system for STR analysis of single source samples. Forensic Sci. Int. Genet. 14, 76–85.
- LaRue, B. et al. (2014) An evaluation of the RapidHIT® system for reliably genotyping reference samples. Forensic Sci. Int. Genet. 13, 104–111.
- McLaren, R.S. et al. (2014) Developmental validation of the PowerPlex® ESI 16/17 Fast and PowerPlex® ESX 16/17 Fast Systems. Forensic Sci. Int. Genet. 13, 195–205.
- Bourdeau, J.M. et al. (2013) Rapid amplification of the European loci in a miniSTR format. Profiles in DNA [cited: 2015, March, 12]. Available from: http://www.promega.com/resources/profiles-in-dna/2014/the-powerquant-system-a-new-quantification-assay-for-determining-dna-concentration-and-quality/ .
- AmpFlSTR® NGM SElect™ PCR Amplification Kit User Guide, Applied Biosystems.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Kutranov, S. et al. Rapid Processing of Casework and Reference DNA Samples Using Standard Protocols. [Internet] 2015. [cited: year, month, date]. Available from: https://www.promega.com/resources/profiles-in-dna/2015/rapid-processing-of-casework-and-reference-dna-samples-using-standard-protocols/
American Medical Association, Manual of Style, 10th edition, 2007
Kutranov, S. et al. Rapid Processing of Casework and Reference DNA Samples Using Standard Protocols. Promega Corporation Web site. https://www.promega.com/resources/profiles-in-dna/2015/rapid-processing-of-casework-and-reference-dna-samples-using-standard-protocols/ Updated 2015. Accessed Month Day, Year.
Contribution of an article to Profiles in DNA does not constitute an endorsement of Promega products.
PowerPlex is a registered trademark of Promega Corporation. SwabSolution is a trademark of Promega Corporation.
AmpFlSTR and POP-4 are registered trademarks of Life Technologies. Applied Biosystems, GeneMapper, Quantifiler and Veriti are registered trademarks of Applied Biosystems. Arktik is a trademark of Finnzymes Oy or its affiliates. FTA is a registered trademark of Flinders Technologies, Pty, Ltd., and is licensed to Whatman. GeneAmp is a registered trademark of Roche Molecular Systems, Inc. NGM SElect is a trademark of Life Technologies. OmniSwab is a trademark of Whatman. QIAsymphony is a registered trademark of QIAGEN GmbH. RapidHIT is a registered trademark of IntegenX, Inc.